Search results for #Scanpy
Collaboration in #scRNA data analysis doesn’t have to be challenging. Explore how C-DIAM can help by letting bioinformatics teams upload and share outputs from #Seurat or #Scanpy with bench scientists for interactive viz. Read our blog for more details: pythiabio.com/post/share-and…
Experience the magic of Tahoe-100M thanks to @fabian_theis's group and #Scanpy.
Experience the magic of Tahoe-100M thanks to @fabian_theis's group and #Scanpy.
Interested in exploring @vevo_ai’s massive Tahoe-100M single-cell perturbation dataset? #Scanpy now supports out-of-core analysis, powered by Dask—allowing you to seamlessly work with a single huge AnnData object without loading everything into memory. 🔗 theislab.github.io/vevo_Tahoe_100…
Chrysalis can be easily integrated into any workflow based on #Scanpy and works seamlessly with multi-sample datasets. This amazing work was led by @Rockdeme. A huge thanks to Jelica Vasiljevic, Kerstin Hahn and @RottenbergSven for their invaluable contributions.
Hopefully this should solve the fight between #Seurat and #scanpy : easySCF: A Tool for Enhancing Interoperability Between R and Python for Efficient Single-Cell Data Analysis | Bioinformatics | Oxford Academic academic.oup.com/bioinformatics…
Chrysalis can be easily integrated into any workflow based on #Scanpy and works seamlessly with multi-sample datasets. A huge thanks again to Jelica Vasiljevic, Kerstin Hahn, @RottenbergSven, and @alvaldeolivas for their invaluable contributions. 🧵 (6/7)
scATAcat is available as a Python package (github.com/aybugealtay/sc…), compatible with #scanpy & #anndata, and comes with a tutorial available at scatacat.readthedocs.io/en/latest/inde…
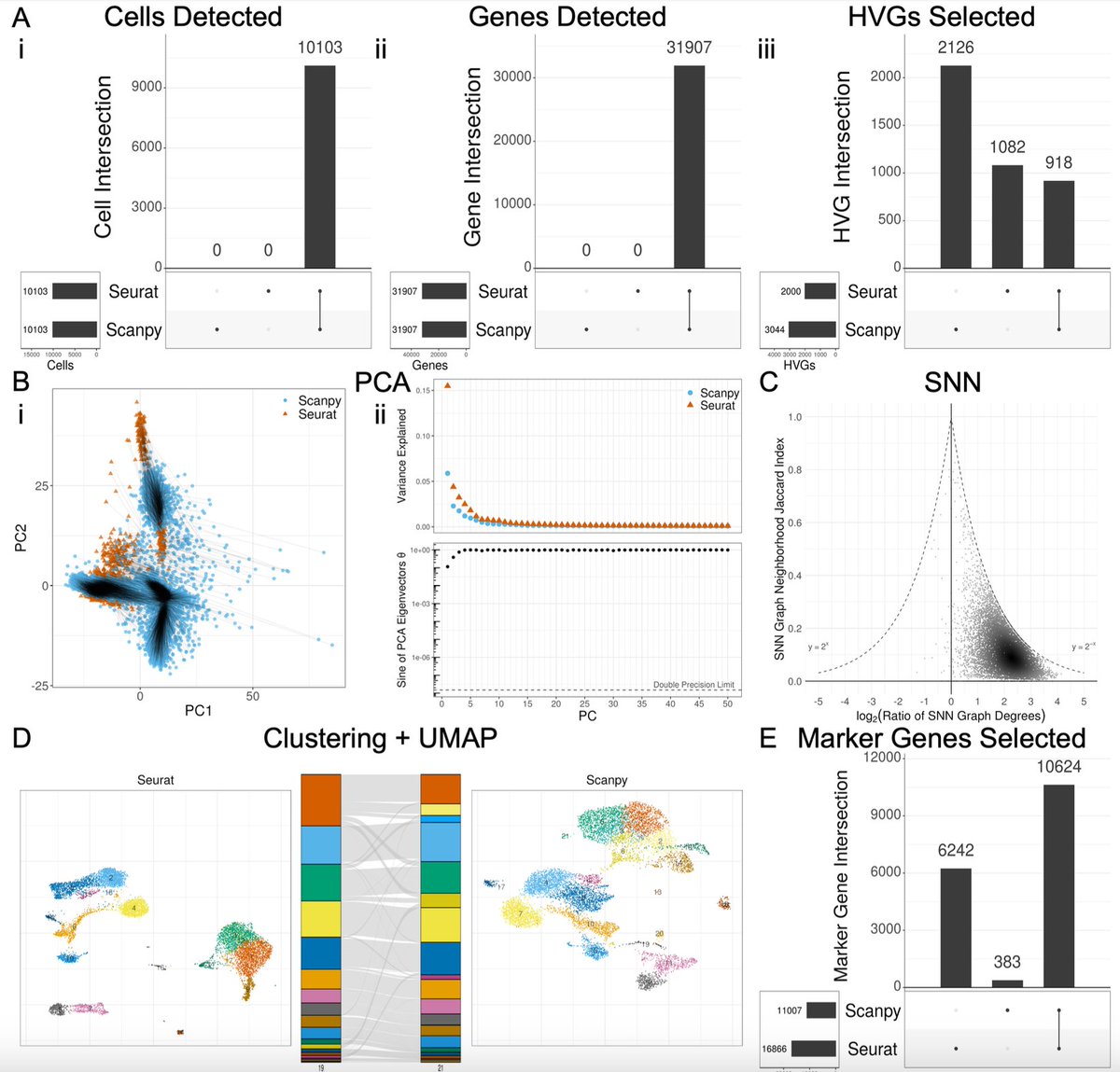
Probably one of the most comprehensive scRNA seq popular softwares ( @satijalab #seurat & @scverse_team #scanpy ) most comprehensive comparison. Loved it.
Probably one of the most comprehensive scRNA seq popular softwares ( @satijalab #seurat & @scverse_team #scanpy ) most comprehensive comparison. Loved it.
If you are doing single cell RNA sequencing, you must have thoughts on Trajectory analysis. There are multiple tools on the github, but which one you used and why ? #seurat #scanpy #Bioinformatics #scRNA
Chrysalis can be easily integrated into any workflow based on #scanpy and works on multi-sample datasets. Huge thanks to Jelica Vasiljevic, Kerstin Hahn, @RottenbergSven, and @alvaldeolivas for their invaluable contributions. 🧵 (5/6)
Authors have used @scverse_team #scanpy for gene expression pre-processing , Episcanpy for chromatin accessibility ,Batch integration and scMultiomics integration was performed using scib.metrices ! /2
This package extends the Infinity Flow protocol to allow for more complex configurations. Data is held in AnnData Python objects to enable seamless integration with popular single-cell omics tools, such as #scanpy and #pytometry. (3/7)
This package extends the Infinity Flow protocol to allow for more complex configurations. Data is held in AnnData Python objects to enable seamless integration with popular single-cell omics tools, such as #scanpy and #pytometry. (2/6)
I've been exploring #GPT models' capabilities in #singlecell analysis for a while now, and they've all struggled with #scanpy. Although #ChatGPT-3.5 showed promise, it fell short in grasping the complexities of single cell RNA-seq data. Enter #GPT4. 👇
I've been exploring #GPT models' capabilities in #singlecell analysis for a while now, and they've all struggled with #scanpy. Although #ChatGPT-3.5 showed promise, it fell short in grasping the complexities of single cell RNA-seq data. Enter #GPT4. 👇

ScanpyRook @ScanpyR63549
1 Followers 3 Following
ARCESE @scanpy
0 Followers 0 Following
scanpy @scanpy1
0 Followers 0 Following
ScanPy @py_scan
0 Followers 1 Following
Alex Wolf @falexwolf
2K Followers 375 Following Open data infra for biology @laminlabs. Previously, created Scanpy & led build-up of Cellarity's compute platform.